Millard Lab - Projects
We are always interested in recruiting enthusiastic scientists and welcome new ideas and techniques. Please contact Sean to enquire about Honours, Masters, PhD or Postdoctoral positions in the lab. Below is a list of ongoing projects.
Dscam2-related projects
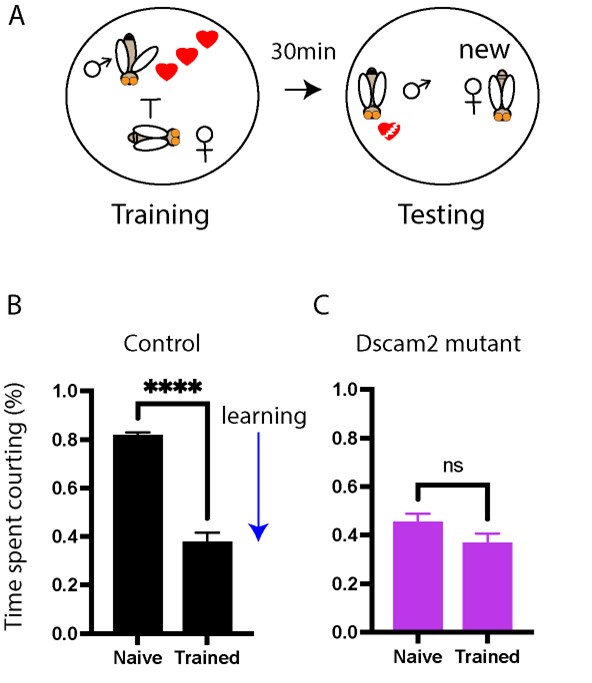
The role of Dscam2 in learning and memory
In collaboration with Krystyna Keleman at QBI.
We are using a courtship conditioning assay to assess short-term memory in flies. Dscam2 mutants have a memory defect and we aim to figure out the role that it plays in this proces
Molecular mechanisms of Dscam2 repulsion
We have found that Dscam2 homophilic binding signals through the PI3K pathway to induce repulsion. We have an in vivo assay set up to measure this and we are using it to uncover the molecular players involved and how homophilic binding leads to their recruitment.
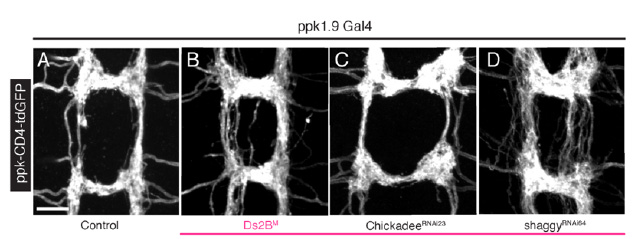
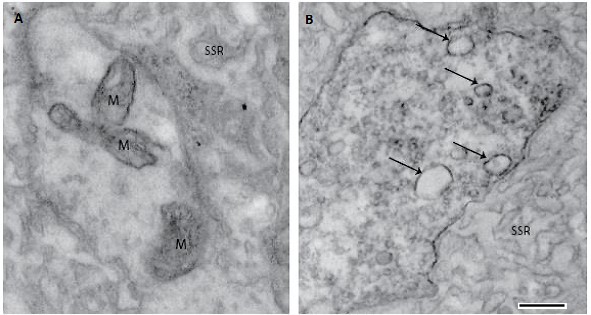
How does Dscam2 inhibit synaptic strength at the larval NMJ?
Dscam2 suppresses synaptic strength in motor neurons and we are investigating the cellular mechanisms involved. Synaptic vesicle recycling appears to be critical.
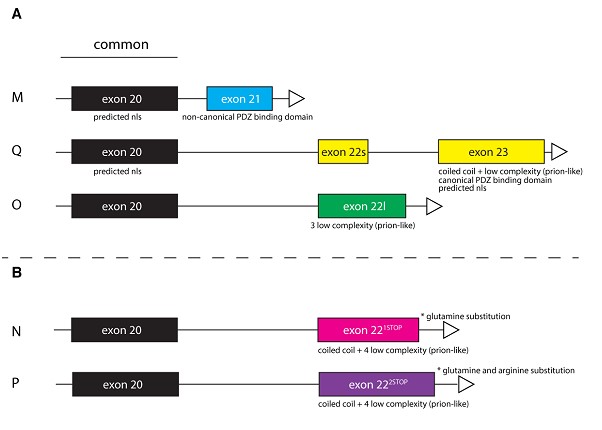
Do different Dscam2 cytoplasmic isoforms have different functions?
Dscam2 has 5 alternatively spliced cytoplasmic isoforms that have very different protein compositions. We are investigating whether they have different functions at the larval NMJ.
ALS project
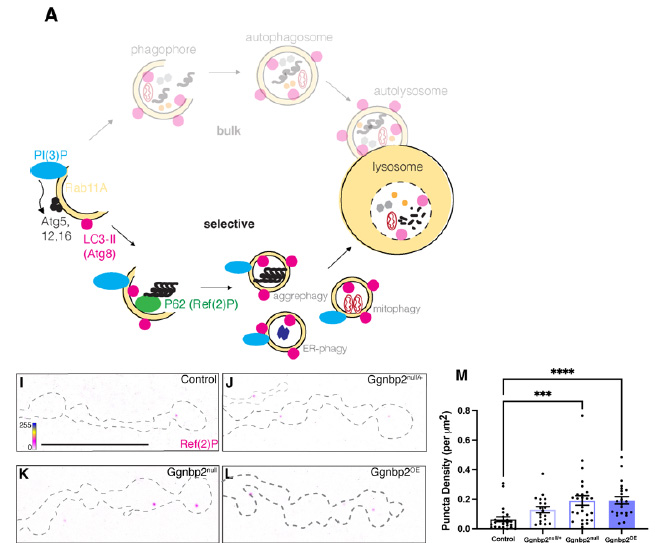
How the ALS risk factor, GGNBP2, contributes to autophagy
In collaboration with Naomi Wray’s group at IMB.
We have been studying how the gene, GGNBP2, contributes to ALS pathology using the fly. In humans, increased expression of this gene is correlated with increased risk of ALS. We have shown that it is required for synaptic development in fly motor neurons and that it is involved in autophagy, a process commonly defective in ALS.
Mitophagy project
Mechanisms of Fbxl4-mediated mitophagy in Drosophila
In collaboration with Julia Pagan’s lab at SBMS.
Mitophagy is the selective degradation of damaged mitochondria using autophagic machinery. The Pagan lab has identified a ubiquitin ligase (FbxL4) that limits mitophagy by degrading receptors on mitochondria required for this process. We are trying to understand the mechanisms involved using Drosophila.
