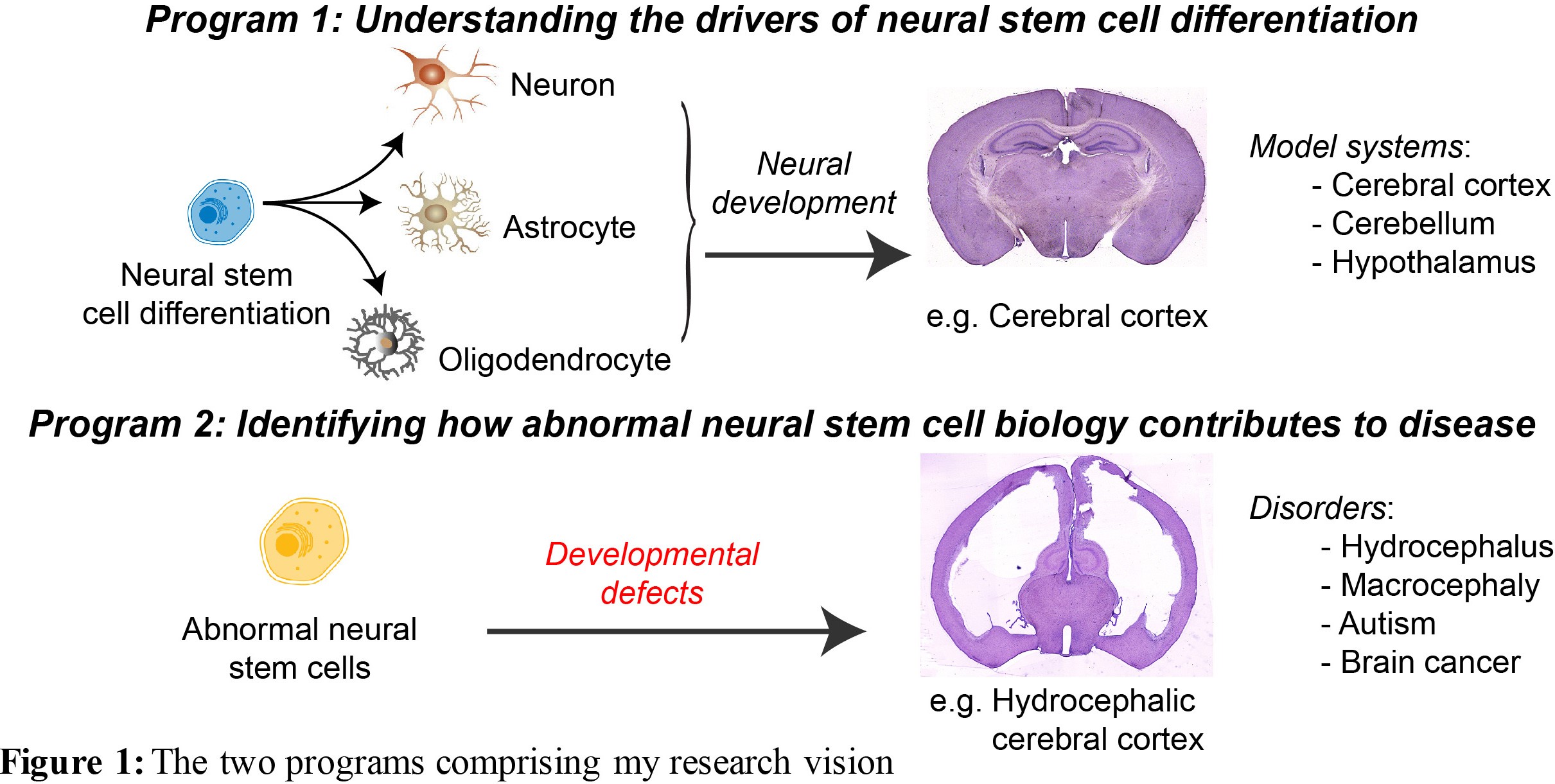
Research directions
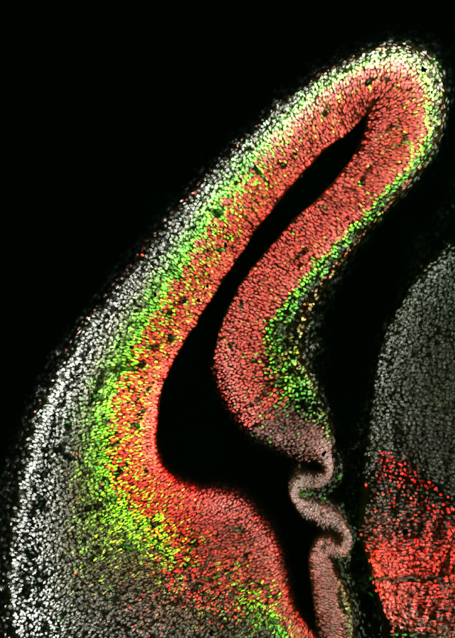
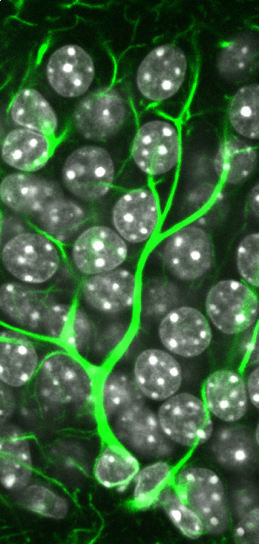
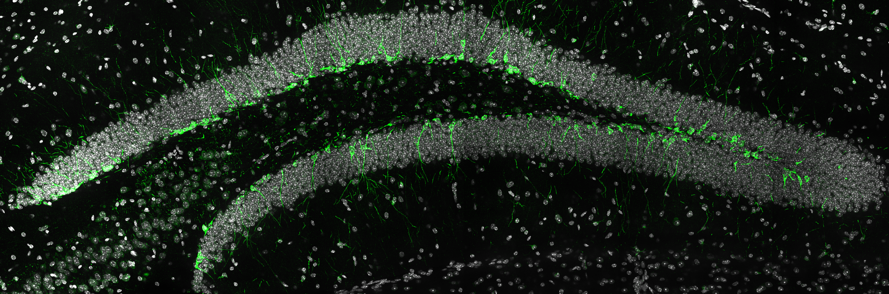
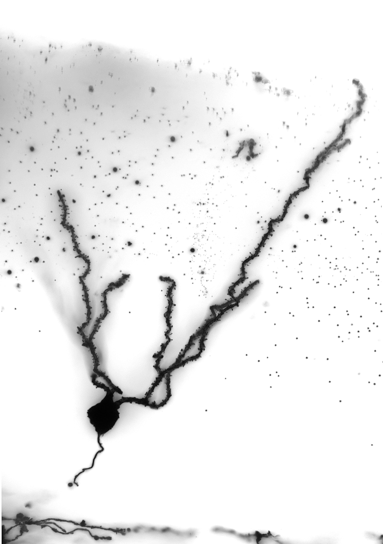
The human brain is an incredibly complex organ, consisting of over 100 billion neurons, and even more glial cells. Further adding to this complexity is the fact that there are a wide variety of distinct neuronal subpopulations within the brain, each with different morphological characteristics, neurochemical properties and patterns of connectivity. Amazingly, nearly all of the cells within the brain are derived from a relatively small population of neural stem cells (NSCs) that proliferate, then differentiate, during embryogenesis. Understanding how NSC biology is coordinated, both spatially and temporally, to generate the mature brain remains one of the great challenges in biology. My vision is to reveal the mechanisms that control NSC differentiation within the developing brain, and to apply this knowledge to understand disease processes caused by abnormal NSC differentiation (Figure 1).
I have made a number of significant contributions to understanding how NSC differentiation is coordinated during neural development since starting my own group in late 2010. This work, which was supported by competitive fellowship (NHMRC Career Development Fellowship 2009-2012; ARC Future Fellowship 2013-2017) and grant funding (three NHMRC project grants as CIA; two ARC Discovery Projects as sole CI), has helped to elucidate the fundamental mechanisms underpinning neurogenesis within the neocortex, hippocampus and cerebellum. I have also defined critical molecular controllers of NSC quiescence, a cellular state that ensures the longevity of adult NSCs, as well as describing the behavioural consequences of aberrant adult neurogenesis. Finally, I have provided new insights into how abnormal stem cell biology can contribute to a range of neurodevelopmental disorders, as well as cancers of the brain and skin. The significance of my findings has been recognised by multiple awards for research excellence, from both national (e.g. 2018 Emerging Leader Award, Australian and New Zealand Society for Cell and Developmental Biology; 2010 AW Campbell Award, Australasian Neuroscience Society) and international agencies (2015 Innovator Award, Hydrocephalus Association; 2010 CJ Herrick Award, American Association for Anatomists). I now am in an ideal position to address aspects of two key questions in the field, namely, what are the transcriptomic and epigenomic factors that control the differentiation of NSCs during brain development, and how do deficits in this process contribute to disease?
For my full publication track record, please visit my ORCID site
- Shim WJ, et al. Conserved Epigenetic Regulatory Logic Infers Genes Governing Cell Identity. Cell Syst. 2020;11(6):625-39 e13.
- Yoon S, et al. Usp9X Controls Ankyrin-Repeat Domain Protein Homeostasis during Dendritic Spine Development. Neuron. 2020;105(3):506-21 e7.
- Kasherman MA, et al. Abnormal Behavior and Cortical Connectivity Deficits in Mice Lacking Usp9x. Cereb Cortex. 2021;31(3):1763-75.
- Kojic M, et al. Elp2 mutations perturb the epitranscriptome and lead to a complex neurodevelopmental phenotype. Nat Commun. 2021;12(1):2678.
- Weissleder C, et al. Reduced adult neurogenesis is associated with increased macrophages in the subependymal zone in schizophrenia. Mol Psychiatry. 2021;26(11):6880-95.
- Davila RA, et al. Deletion of NFIX results in defective progression through meiosis within the mouse testisdagger. Biol Reprod. 2022;106(6):1191-205.
- Gaik M, et al. Functional divergence of the two Elongator subcomplexes during neurodevelopment. EMBO Mol Med. 2022;14(7):e15608.
- Yaghmaeian Salmani B, et al. Selective requirement for polycomb repressor complex 2 in the generation of specific hypothalamic neuronal subtypes. Development. 2022;149(5).
- Mitchell B, Thor S, Piper M. Cellular and molecular functions of SETD2 in the central nervous system. J Cell Sci. 2023;136(21).
- Currey L, et al. Polycomb repressive complex 2 is critical for mouse cortical glutamatergic neuron development. Cereb Cortex. 2024;34(7).
Group Head
Staff
PhD and Honours students
- Ben Mitchel, PhD student
- Cooper Atterton, PhD student
- Douglas Kirkland, Honours student
October 2025
Well done to Hallie, who handed in her Honours thesis in early October. An amazing effort!
Well done also to Cooper, as well as Hallie, Ben, Bee and Laura, as their paper on how SETD2 regulates brain development was recently published.
Laura recently started a new position with Dr Jacky Suen; well done Laura!
June 2025
Well done to Cooper, who smashed his Progress Review #1 this month.
Laura had her review on the underlying causes of macrocephaly in patients with ASD published this month. This appears in Frontiers in Neuroscience. Check it out!
May 2025
Congratulations to Cooper, who just had his review on overgrowth disorders published in Disease Models and Mechanisms. This paper was a collaborative effort with two patient-focussed advocacy groups, the Malan Syndrome Foundation and the Tatton-Brown-Rahman community, as well as with collaborators from Murdoch University.
Well done to Hallie, who submitted her Honours proposal this month
A big well done to Laura, Tracey and Sandy, who had their review on the molecular mechanisms underpinning brain overgrowth in ASD recently accepted to Frontiers in Neuroscience. This was a massive effort, and this review fills a big gap in the literature.
March 2025
A big welcome to Hallie, who has joined us for Honours this year
Congrats to Ben, whose lovely article on how to perform Golgi-Cox staining on embryonic brain tissue has just been published.
Cooper’s paper on the role of TEAD1 in the developing cerebellum is now available online. Check it out!
February 2025
Well done to Cooper, whose paper on the role of TEAD1 in cerebellar development was just accepted to Brain Structure and Function
We also had a collaborative paper with the Perlmann lab recently accepted to Development
A collaborative study with the Day lab was recently published in Nature Communications.
The Piper lab were fortunate to collaborate with the Rawashdeh lab on a paper on the role of melatonin in the timing of sleep onset. This paper is now published in npj Biological Timing and Sleep.
Congrats also to Laura, whose first author paper describing some of her sequencing studies was recently published.
The lab was lucky to host a summer student, Bee Miebusch, over the break. Bee was a great addition to the lab!
November 2024
Well done to Laura, whose short research note was recently accepted to BMC Research Notes.
A huge congratulations to Lucy, who submitted her Honours thesis. A massive effort.
And finally, well done to Sandy, whose lovely paper mapping the expression of TEAD1 in the developing mouse brain was recently published in Gene Expression Patterns.
September 2024
This was a good month for manuscript submissions, with Cooper, Sandy and Laura all submitting papers. We also had collaborative papers from the Boden and Perlmann labs submitted this month. Fingers crossed!
We also had a collaborative manuscript with the Rawashdeh lab accepted to Biological Timing and Sleep this month.
July 2024.
Well done to Laura, whose paper was published in full in Cerebral Cortex.
We also had a collaborative paper with the Day lab (QIMRB) accepted in Nat Comm this month.
The lab was also lucky enough to have Anna join us in July for a UQ Winter Scholarship.
June 2024
Laura’s paper has been accepted to Cerebral Cortex. Well done Laura! Thank you also to the past and present lab members (Ben, Sandy, Danyon and Lachlan) and our wonderful collaborators for their contributions to this great piece of work.
Well done to Lucy, who got excellent marks for her Honours proposal and seminar.
Welcome to Anna, who joined the lab for a Winter Scholarship.
Congratulations to Mikki and Justin, both of who received Fist Class Honour for their projects. Well done!
May 2024
Well done to Mikki and Justin on completing their Honours years. Both did very well, and the year culminated in Mikki winning the best oral in the Honours seminar presentations!

Lucy also received her marks back for her Honours proposal seminar, and did very well indeed! Well done!
April 2024
Ben passed his first Progress Review with flying colours. Well done mate!
Lucy also did extremely well in her Honours proposal seminar, and she is poised for a big year ahead
Well done to both Mikki and Justin for completing their Honours projects. A great effort!
Welcome to Cooper, who joined us as a PhD scholar this research quarter.
Well done also to Raul who submitted his PhD thesis this month. Almost there now!!
February 2024
A big thank you to Hallie Naumann, who worked with us over the summer. And a big hello to Linda Luo, who will be in the lab for semester 1.
Ben’s paper has now been published in Neuroscience Letters. Check it out!
If you would like to make a tax deductible donation to neural stem cell research, please contact med.advancement@uq.edu.au. Thank you for your support.
Find out more about our diverse range of research interests.